Global genomic studies reveal high diversity of drug-resistant Escherichia coli in diabetic foot infections.
A multinational collaborative study has for the first time systematically analyzed the genomic characteristics of Escherichia coli from diabetic foot ulcer samples from multiple countries around the world, providing key scientific evidence for understanding the refractory nature of infection and clinical prognosis.
A groundbreaking international study, jointly led by King's College London and the University of Westminster, has revealed astonishing genetic diversity in E. coli detected in diabetic foot infections, with many bacteria carrying multiple antibiotic resistance genes and pathogenic factors. This groundbreaking research provides the first comprehensive genomic analysis of E. coli originating from multiple continents in diabetic foot infections, and the findings have been published in the prestigious journal *Microbiology Spectrum*.
Diabetic foot infection is one of the most serious complications of diabetes and a leading cause of non-traumatic lower limb amputations worldwide. Although the complexity of this type of chronic wound infection has long been recognized in clinical practice, the scientific community's understanding of the specific characteristics and harmful mechanisms of the key pathogens involved, especially Escherichia coli, remains limited.
To fill this knowledge gap, the research team performed whole-genome sequencing and in-depth analysis on 42 Escherichia coli strains isolated from patient samples from ten countries, including Nigeria, the UK, Ghana, Sweden, Malaysia, China, South Korea, Brazil, India, and the US. By comparing the genetic differences among different strains, researchers were able to explore the bacterial biological characteristics associated with diabetic foot disease on a global scale.
Key findings include:
These infecting strains belong to numerous different genetic lineages and do not originate from a single culprit clone. This indicates that the unique environment of diabetic foot ulcers can be successfully adapted to and colonized by multiple independently evolving E. coli lineages.
Genomic analysis detected abundant antibiotic resistance genes. Notably, approximately 8% of the strains were identified as multidrug-resistant or extensively drug-resistant, meaning they are insensitive to most, if not almost all, commonly used antibiotics in clinical practice.
The strain carries a variety of virulence factors related to pathogenicity, such as genes that help the bacteria adhere to host tissues or evade immune attacks. These "arsenals" may collectively contribute to the severity and prolongation of the infection.
“This study essentially explains why some diabetic foot infections are exceptionally stubborn and prone to rapid deterioration,” noted Dr. Vincenzo Torraca, senior author of the study and lecturer in infectious diseases at King’s College London. “The genomic data clearly show that we are not facing a single enemy, but a diverse ‘bacterial army’ equipped with multiple drug resistance mechanisms and pathogenic tools.”
This study provides a key direction for improving clinical diagnosis and treatment strategies for diabetic foot infections. By identifying regionally dominant drug-resistant strains and their characteristics, it is hoped that faster and more precise targeted therapy can be achieved in the future, thereby effectively shortening the infection cycle and reducing hospitalization rates and the risk of amputation.
The research team stated that their next step will focus on further exploring the specific mechanisms of action of identified virulence factors within the context of diabetic foot infection. This will open new avenues for developing novel treatments and intervention strategies.
About this study: This was a multinational collaborative project aimed at revealing the evolution and pathogenicity of pathogenic *Escherichia coli* in diabetic foot infections using cutting-edge genomics technologies. All analyses were based on pure cultures of bacteria directly isolated from diabetic foot ulcers in patients.
source:
King's College London

 English
English عربى
عربى Español
Español русский
русский 中文简体
中文简体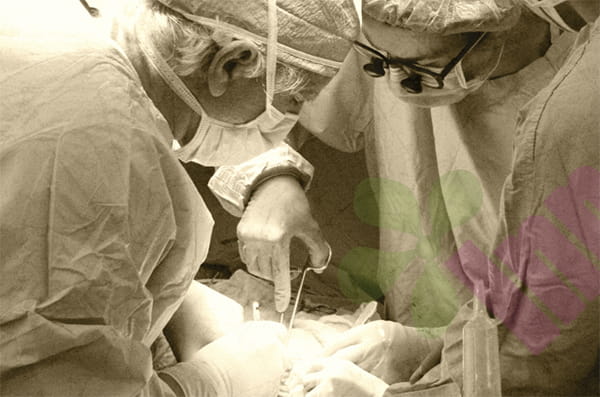









.jpg.png)
.jpg.png)

.jpg.png)


